Maize is one of the three most important food staples globally. However, in spite of this global importance and highly diverse genetic base, the germplasm pool typically used by maize breeders to develop varieties globally is relatively narrow, considering the breadth of diversity in the primary gene pool.

Antonio Hernández and Oscar Osorio measuring field harvest weight at CIMMYT’s Agua Fría station. Photo: CIMMYT.
Maize genebanks of the world conserve many tens of thousands of landraces, each representing a population of genetically heterogeneous individuals. These were, and, in regions of some countries like Mexico, are still used by smallholder farmers. In addition to landraces, the banks also hold many hundreds of accessions of maize ancestors and wild relatives (teocinte and Tripsacum species). Together, these collections represent the genetic heritage of maize, potentially harboring many undiscovered genes, alleles and haplotypes that have yet to be harnessed for maize improvement.
In the Maize component of SeeD we systematically explore the genetic richness of these collections to enable the targeted broadening of the genetic base of maize breeding programs. We are working towards four interrelated objectives, each comprising a group of well-defined outputs:
- Objective 1: Build a molecular atlas for maize. We have developed a sequence based genotyping method that uses leaf bulks to simultaneously quantify genetic distances amongst, and diversity parameters within landrace populations (accessions) of genetically heterogeneous individuals. This move from single-plant to population-level genetic fingerprints (comprising both SNP and PAV polymorphisms) makes it possible to systematically characterize up to 40,000 landraces, advanced lines and wild relatives. The resulting genotypic data, along with links to existing passport and characterization/evaluation data, will be represented in a searchable ‘Maize Molecular Atlas’, available through the Germinate platform via this website.
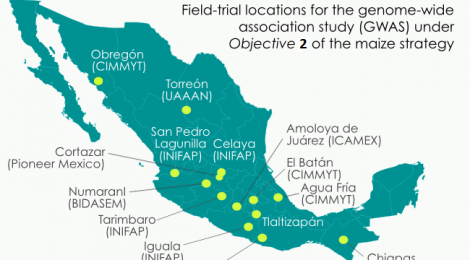
- Objective 2: Identify alleles for stress-tolerance traits and traditional uses. Many maize breeding targets are genetically complex. Under this objective, SeeD aims to identify alleles and haplotypes from landraces, which control traits such as heat and drought tolerance. As part of this effort SeeD has developed a population of more than 4,000 testcross progenies derived from accessions of CIMMYT’s core collection. Only one individual per accession was crossed, based on the assumption that haplotypes are likely to be replicated across accessions. Project participants have been phenotyping this population across multiple experimental stations in the different agroecological zones where maize is grown in Mexico. The testcross parents have been genome-profiled at ultrahigh density using DArTseq methodology. The trial data to date have been co-analyzed with the genome-profile data to perform a large genomic prediction/genome-wide association study (GWAS) to identify high-value ‘donor accessions’ and beneficial small-effect alleles/haplotypes. In addition to this effort SeeD has been using derived GIS (Geographic Information System) data obtained from analysis of collecting sites for both the CIMMYT germplasm bank collection and a recent collection funded by the National Commission for the Knowledge and Use of Biodiversity (CONABIO) to select accessions based on environmental or usage data and screen for beneficial alleles for selected traits. The screening involves (a) the per se characterization of accessions e.g. collected from extreme environments, heat and drought tolerance; identified for specific culinary/nutritional properties; blue maize, and (b) the evaluation of well performing accessions in testcross background.
- Objective 3: Develop maize ‘bridging germplasm’. The final objective harnesses and combines outputs of the first three to develop new early generation maize lines in breeder friendly backgrounds with introgressed alleles of beneficial effect. This bridging or ‘pre-breeding germplasm’ will be made available to breeders, along with associated information such as agronomic performance in stress and optimal conditions, marker-trait associations and genomic estimates of breeding values to facilitate better selection and use of germplasm for trait mobilization within breeding programs.

Terry Molnar, maize breeder with SeeD, and Enrique Rodriguez, field research technician with SeeD, evaluate bridging germplasm for resistance to tar spot complex. Photo: J. Johnson/CIMMYT.
To stimulate innovation and crowdsourcing-based data mining, all maize datasets and attribution information will be made publicly available via the Maize Molecular Atlas no later than 24 months after their generation (so that those who generated the data have an opportunity to draft scientific publications to add value to the data).
For more information please contact us at: seed@masagro.org